Our research
Membrane trafficking
Eukaryotic cells are compartmentalised which means that each intracellular membrane has a distinct set of functions and associated molecules. These compartments are interconnected by membrane trafficking pathways that allow the exchange of proteins and lipids between them.
Clathrin-mediated endocytosis is a major pathway for the internalisation of cell surface proteins and lipids. Projects in the lab are to understand how this mode of endocytosis works at the molecular level. For example, we have engineered a system to allow us to trigger clathrin-mediated endocytosis on-demand. We have used this system to demonstrate the minimal machinery needed to form a clathrin-coated vesicle and with this knowledge, we could make clathrin-coated pits on membranes that don’t normally have them (link).
During intracellular traffic, we idientified a new type of transport vesicle that we call INVs (intracellular nanovesicles). These vesicles are small in size (30-40 nm diameter) and carry proteins on the anterograde and recycling pathways in the cell (link). We are working to define the cargos they contain as well as the molecular machinery that drives their formation and movement.
Cell division
Chromosome segregation is probably the most important moment in a cell’s life. Any error in sharing the duplicated genome equally to the two daughter cells results in aneuploidy. We know that most solid tumours are aneuploid and it is likely that this form of genetic instability underlies the malignancy.
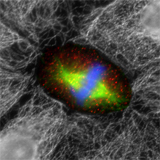
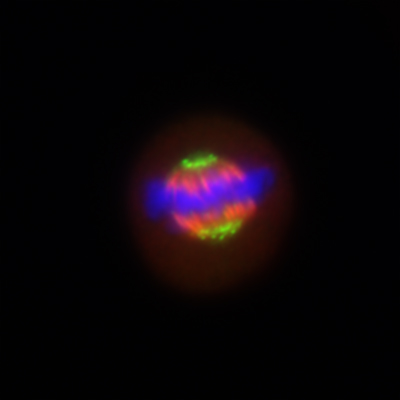
The cell builds a tiny machine – the mitotic spindle – to achieve accurate chromosome segregation in mitosis. The mitotic spindle comprises microtubules and hundreds of associated proteins as well as the chromosomes themselves.
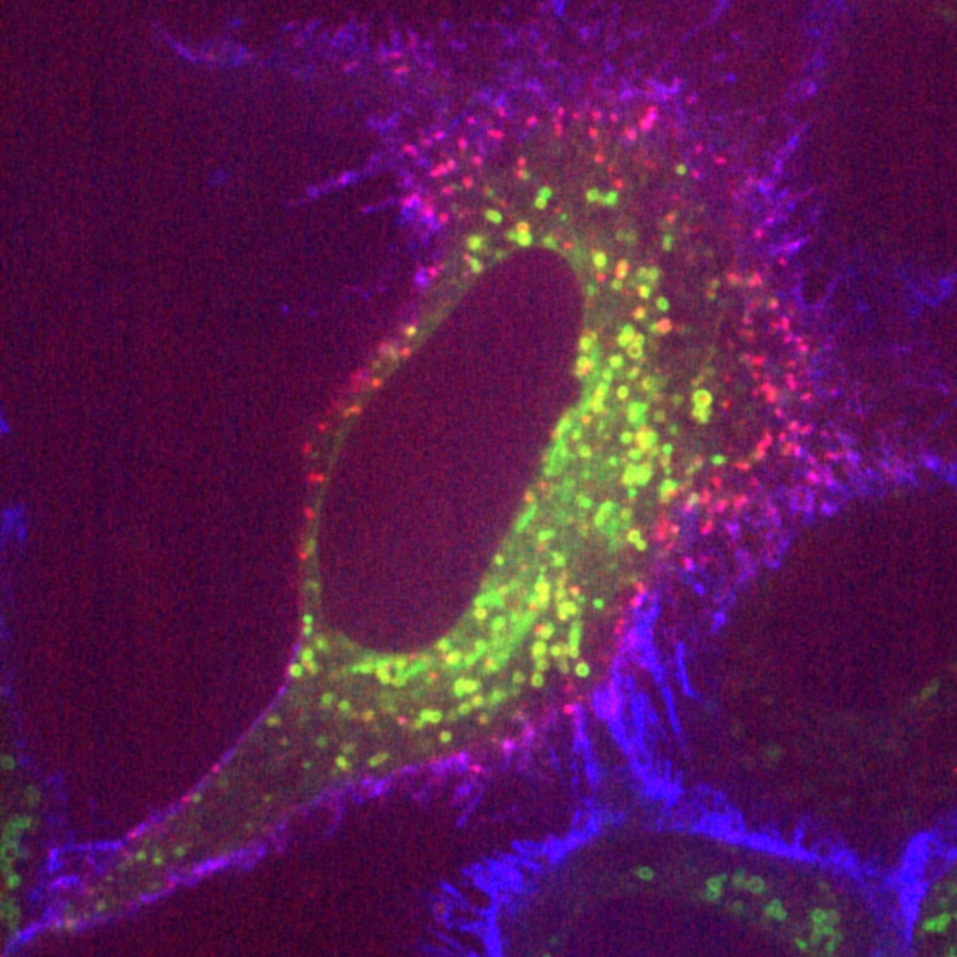
To understand how the microtubules in the spindle are organised, we have focussed on crosslinking proteins which stabilise fibres of microtubules. We discovered a complex of clathrin, ch-TOG, TACC3 and GTSE1 that forms bridges between microtubules (here). Recent projects have focused on trying to inhibit the formation of the complex with the goal of interfering with the spindle and causing cell death.
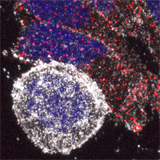
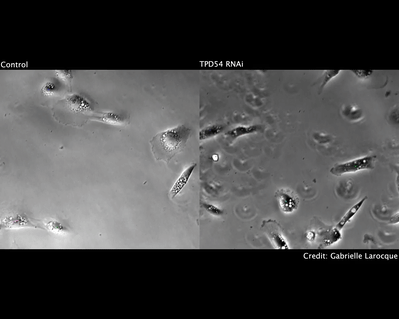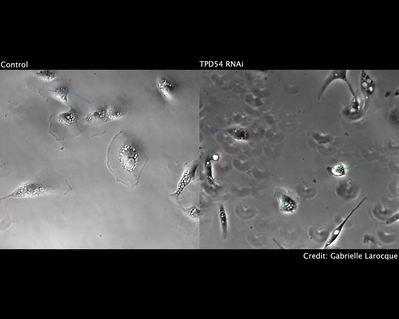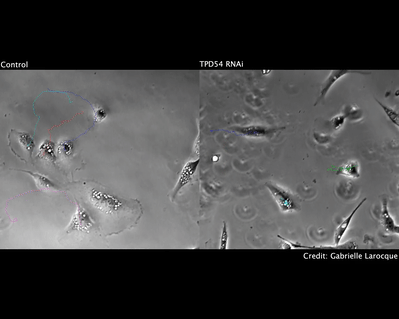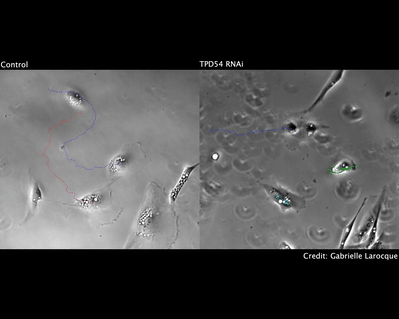
We have also investigated how chromosome mis-segregation occurs in cells. When chromosomes lose attachment to the spindle, they are at risk from becoming ensheathed in endoplasmic reticulum (ER) membranes (link). How these interactions occur and how they are resolved is a current focus of the lab.

Genetic tools for cell biology
If we take cell biological systems and re-engineer them, we can answer questions about how these systems work or use these tools to uncover new cell biology. We call the approach of re-engineering cell biologucal systems, “remixing” cell biology.
Some examples from our lab include:
- FerriTag - a re-engineered ferritin particle that can label proteins for light and electron microscopy.
- Hot-wired endocytosis - a system to trigger clathrin-mediated endocytosis on-demand
- LaBeRling - highlight ER-membrane contact sites on-demand
We wrote an Cell Science at-a-glance paper to describe these type of tools (link).